IT Tools

EspINA
Since synapses are key elements in the structure of nervous circuits, understanding their location, size and proportion between the two different types is extraordinarily important in terms of function.
In this way, EspINA tool automatically performs segmentation and 3D volume reconstruction of synapses in the brain, helping the user to examine large tissue volumes and interactively validate the results provided by the software.
Morales J, Alonso-Nanclares L, Rodríguez J-R, DeFelipe J, Rodríguez Á and Merchán-Pérez Á (2011) ESPINA: a tool for the automated segmentation and counting of synapses in large stacks of electron microscopy images. Front. Neuroanat. 5:18. doi: 10.3389/fnana.2011.00018
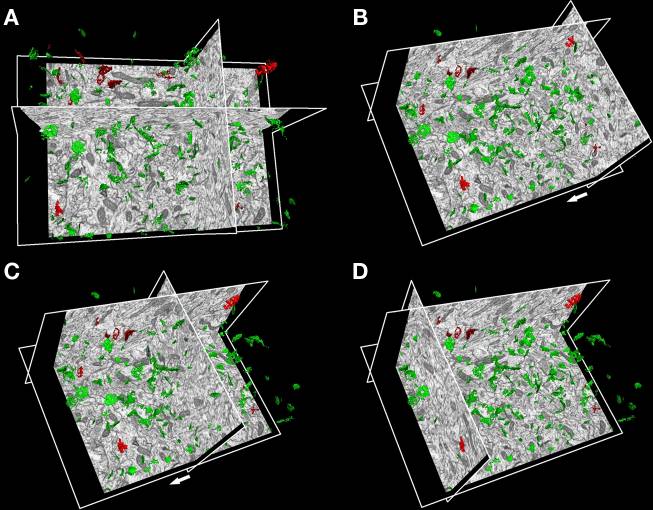
EspINA can display multiple spatially or temporally related images. These image sets are called stacks. The images that make up a stack are called sections. All the sections in a stack must be the same size and bit depth. EspINA supports 8-bit images.
EspINA is a memory intensive application used to reconstruct, refine, analyze and visualize structures in the brain. It’s an open source application and it’s functionalities can be expanded by plugins. It’s currently available for Linux (Ubuntu based) and Windows machines with the only requirement of a 64 bits CPU.

Audispine
We present a new method with musical feedback for exploring dendritic spine morphology and distribution patterns in pyramidal neurons. We demonstrate that audio analysis of spiny dendrites with apparently similar morphology may “sound” quite different, revealing anatomical substrates that are not apparent from simple visual inspection.
Pablo Toharia, Juan Morales, Octavio de Juan, Isabel Fernaud, Angel Rodríguez, Javier DeFelipe. Neuroinformatics, January 2014. Musical representation of dendritic spine distribution: a new exploratory tool

Neuronize
This tool presents a new technique for the generation of three-dimensional models for neuronal cells from the morphological information extracted through computed-aided tracing applications. The 3D polygonal meshes that approximate the cell membrane can be generated at different resolution levels, allowing balance to be reached between the complexity and the quality of the final model.
Neuronize implements a novel approach to generate a realistic 3D shape of the soma from the incomplete information stored in the digitally traced neuron using a physical deformation technique.
The addition of a set of spines along the dendrites completes the model, generating a final 3D neuronal cell suitable for its visualization in a wide range of 3D environments.
Brito JP, Mata S, Bayona S, Pastor L, Defelipe J, Benavides-Piccione R (2013). A tool for building realistic neuronal cell morphologies. Front Neuroanat. 2013 Jun 3;7:15. doi: 10.3389/fnana.2013.00015. eCollection 2013.
Neuronize requires Matlab Compiler Runtime 2012b (you can download freely, clicking on here) to be installed on your computer. Versions for Windows 32 and 64 bits platforms are available. Versions for Linux and Mac are under development.
The video may also help you see how to work with NEURONIZE. It shows a common working session:

Synaptic apposition surface (SAS)
We have developed an efficient computational technique to automatically extract the surface from synaptic junctions that have previously been three dimensionally reconstructed from actual tissue samples imaged by automated FIB/SEM.
This technique has been incorporated into EspINA and SAS structures can be computed automatically from reconstructed synapses.
Juan Morales, Angel Rodríguez, José-Rodrigo Rodríguez, Javier DeFelipe and Angel Merchán-Pérez (2013). Characterization and extraction of the synaptic apposition surface for synaptic geometry analysis Front. Neuroanat., 04 July 2013 | doi: 10.3389/fnana.2013.00020
Laboratory Cajal Cortical Circuits
Centro de Tecnología Biomédica, Universidad Politécnica de Madrid
Campus de Montegancedo s/n
Pozuelo de Alarcón 28223 (Madrid) Spain
Tel: +34 910679250


